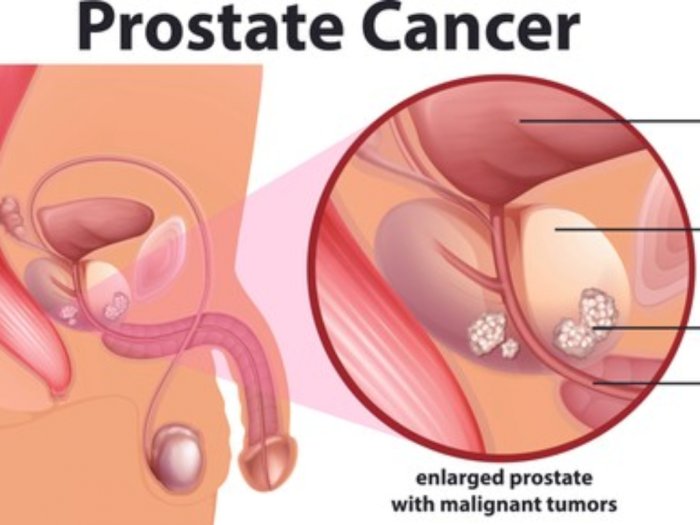
Informasi Tentang Penyakit Kanker Prostat (Prostate Cancer) – Tentang prostat – Prostat adalah kelenjar seukuran kenari yang terletak di belakang pangkal penis, di depan rektum, dan di bawah kandung kemih. Ini mengelilingi uretra, saluran seperti tabung yang membawa urin dan air mani melalui penis.
Fungsi utama prostat adalah membuat cairan mani, cairan dalam air mani yang melindungi, menopang, dan membantu mengangkut sperma. sbobet88 slot
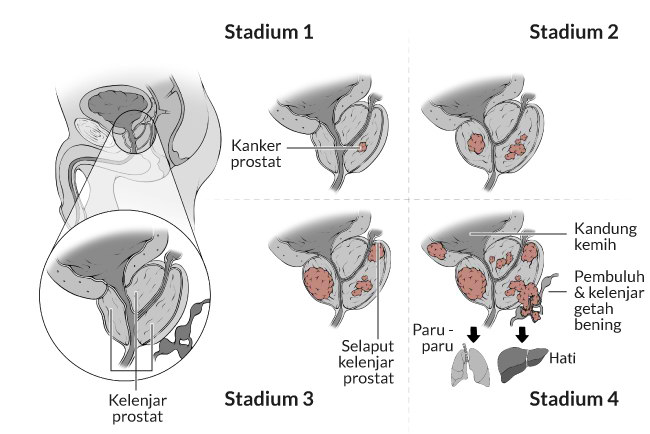
Prostat terus membesar seiring bertambahnya usia. Ini dapat menyebabkan kondisi yang disebut hipertrofi prostat jinak (BPH), yaitu ketika uretra tersumbat. BPH adalah kondisi umum yang terkait dengan bertambahnya usia, dan belum dikaitkan dengan risiko lebih besar terkena kanker prostat.
Tentang kanker prostat
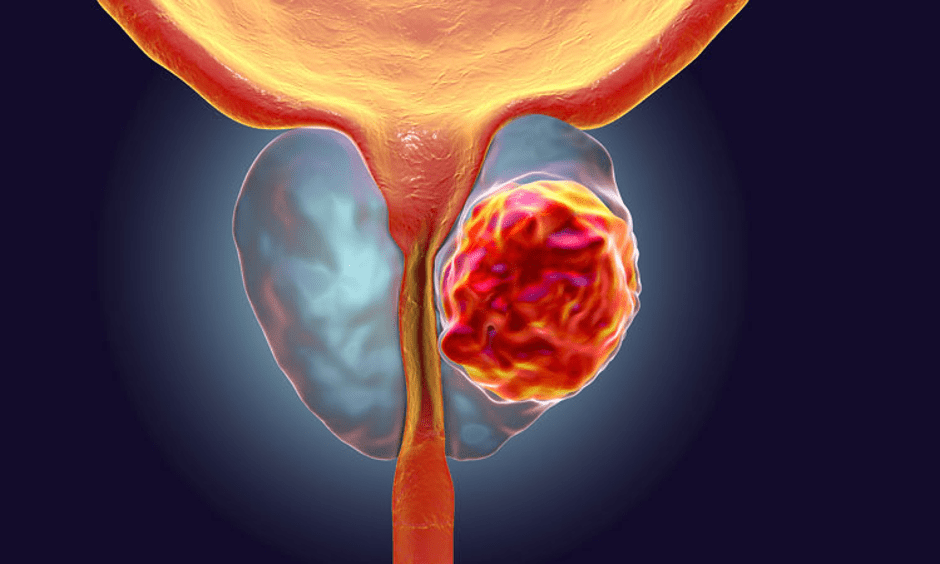
Kanker dimulai ketika sel-sel sehat di prostat berubah dan tumbuh di luar kendali, membentuk tumor. Tumor bisa bersifat kanker atau jinak.
Tumor kanker bersifat ganas, artinya dapat tumbuh dan menyebar ke bagian tubuh lainnya. Tumor jinak berarti tumor dapat tumbuh tetapi tidak akan menyebar.
Kanker prostat agak tidak biasa jika dibandingkan dengan jenis kanker lainnya. Ini karena banyak tumor prostat tidak menyebar dengan cepat ke bagian tubuh lainnya. Beberapa kanker prostat tumbuh sangat lambat dan mungkin tidak menimbulkan gejala atau masalah selama bertahun-tahun atau selamanya.
Bahkan ketika kanker prostat telah menyebar ke bagian lain dari tubuh, seringkali dapat dikelola untuk waktu yang lama. Jadi orang dengan kanker prostat, dan bahkan mereka yang menderita kanker prostat stadium lanjut, dapat hidup dengan kesehatan dan kualitas hidup yang baik selama bertahun-tahun.
Namun, jika kanker tidak dapat dikendalikan dengan baik dengan perawatan yang ada, dapat menyebabkan gejala seperti rasa sakit dan kelelahan dan terkadang dapat menyebabkan kematian.
Bagian penting dari pengelolaan kanker prostat adalah memantau pertumbuhannya dari waktu ke waktu, untuk mengetahui apakah ia tumbuh lambat atau cepat. Berdasarkan pola pertumbuhan, dokter Anda dapat memutuskan pilihan pengobatan terbaik yang tersedia dan kapan harus memberikannya.
Histologi adalah bagaimana sel kanker terlihat di bawah mikroskop. Histologi yang paling umum ditemukan pada kanker prostat disebut adenokarsinoma. Jenis histologis lainnya yang kurang umum termasuk kanker prostat neuroendokrin dan kanker prostat sel kecil.
Varian langka ini cenderung lebih agresif, menghasilkan PSA jauh lebih sedikit, dan menyebar ke luar prostat lebih awal. Baca lebih lanjut tentang tumor neuroendokrin.
Tentang antigen spesifik prostat (PSA)
Antigen spesifik prostat (PSA) adalah protein yang diproduksi oleh sel-sel di kelenjar prostat dan dilepaskan ke dalam aliran darah. Kadar PSA diukur menggunakan tes darah. Meskipun tidak ada yang namanya “PSA normal” untuk siapa pun pada usia tertentu, tingkat PSA yang lebih tinggi dari normal dapat ditemukan pada orang dengan kanker prostat.
Kondisi prostat non-kanker lainnya, seperti BPH (lihat di atas) atau prostatitis juga dapat menyebabkan peningkatan kadar PSA. Prostatitis adalah peradangan atau infeksi pada prostat. Selain itu, beberapa aktivitas seperti ejakulasi dapat meningkatkan kadar PSA untuk sementara.
Ejakulasi harus dihindari sebelum tes PSA untuk menghindari tes peningkatan palsu. Orang harus mendiskusikan dengan dokter perawatan primer mereka pro dan kontra dari pengujian PSA sebelum menggunakannya untuk menyaring kanker prostat.
Ikhtisar pengobatan
Dalam perawatan kanker, berbagai jenis dokter — termasuk ahli onkologi medis, ahli bedah, dan ahli onkologi radiasi — sering bekerja sama untuk membuat rencana perawatan keseluruhan yang dapat menggabungkan berbagai jenis perawatan untuk mengobati kanker.
Ini disebut tim multidisiplin. Tim perawatan kanker mencakup berbagai profesional perawatan kesehatan lainnya, seperti ahli perawatan paliatif, asisten dokter, praktisi perawat, perawat onkologi, pekerja sosial, apoteker, konselor, ahli diet, terapis fisik, dan lain-lain.
Deskripsi jenis perawatan umum yang digunakan untuk kanker prostat tercantum di bawah ini. Rencana perawatan Anda mungkin juga mencakup pengobatan untuk gejala dan efek samping, bagian penting dari perawatan kanker.
Pilihan dan rekomendasi pengobatan tergantung pada beberapa faktor, termasuk jenis dan stadium kanker, kemungkinan efek samping, dan preferensi pasien serta kesehatan secara keseluruhan.
Perawatan kanker dapat mempengaruhi orang dewasa yang lebih tua dengan cara yang berbeda.
Luangkan waktu untuk mempelajari tentang pilihan perawatan Anda dan pastikan untuk mengajukan pertanyaan jika ada sesuatu yang tidak jelas.
Bicarakan dengan dokter Anda tentang tujuan dari setiap perawatan, kemungkinan pengobatan akan berhasil, apa yang dapat Anda harapkan saat menerima perawatan, dan kemungkinan efek samping pengobatan yang berhubungan dengan urin, usus, seksual, dan hormon.
Diskusikan dengan dokter Anda bagaimana pilihan pengobatan dapat mempengaruhi kekambuhan, kelangsungan hidup, dan kualitas hidup. Selain itu, penting untuk mendiskusikan pengalaman dokter Anda dalam mengobati kanker prostat. Jenis pembicaraan ini disebut “pengambilan keputusan bersama”.
Pengambilan keputusan bersama adalah ketika Anda dan dokter Anda bekerja sama untuk memilih perawatan yang sesuai dengan tujuan perawatan Anda. Pengambilan keputusan bersama sangat penting untuk kanker prostat karena ada pilihan pengobatan yang berbeda. Pelajari lebih lanjut tentang membuat keputusan perawatan.
Karena sebagian besar kanker prostat ditemukan pada tahap awal ketika mereka tumbuh perlahan, Anda biasanya tidak perlu terburu-buru untuk membuat keputusan pengobatan.
Selama waktu ini, penting untuk berbicara dengan dokter Anda tentang risiko dan manfaat dari semua pilihan pengobatan Anda dan kapan pengobatan harus dimulai. Diskusi ini juga harus membahas keadaan kanker saat ini, seperti:
- Apakah Anda memiliki gejala atau kadar PSA meningkat dengan cepat
- Apakah kanker telah menyebar ke tulang
- Riwayat kesehatan Anda
- Kualitas hidup Anda
- Fungsi kemih dan seksual Anda saat ini
- Kondisi medis lain yang mungkin Anda miliki